Notes for Understanding Evolution - Chapter 15
Click link to return to Biology
409 Schedule
or back to Chapter 14
or ahead to Chapter 16
General guide on these review questions here
Notes for Chapter 15: Molecular Evolution
Introduction
I. Neutral Theory of Molecular Evolution
RQUE15.1: Do differences observed in the corresponding DNA or amino acid sequences of two organisms tend to be neutral with respect to natural selection or are there likely to be corresponding differences in how they affect the fitness of the two organisms?
II. Cytochrome c
The text is sadly out of date, when it states (UE, p. 159) this about cytochrome c: "No other protein has been so fully analyzed for so many different organisms. This ubiquitous protein has been extracted, purified, and analyzed for more than 40 eukaryotic species."
In truth, sequencing 40 organisms for a 300 base pair stretch of a gene would be a decent semester-long undergraduate research project these days. Actually, cytochrome c is much too short (only about 100 amino acids or 300 bp) to be used for reliable reconstruction of phylogeny. This is why there are actually relatively few cytochrome c sequences compared with some other, more commonly compared, protein sequences.
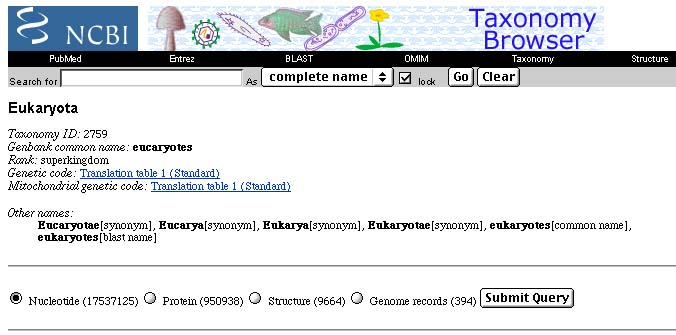
Just for fun, I looked (in July, 2002) in GenBank's Taxonomy Browser to see how many sequences there are for eukaryotes. There were about 17.5 million DNA sequences and nearly a million protein sequences.

Update: As of March, 2003, about eight months later, there are now over 23 million nucleotide sequences (an increase of over 33%) and over 1.1 million protein sequences!
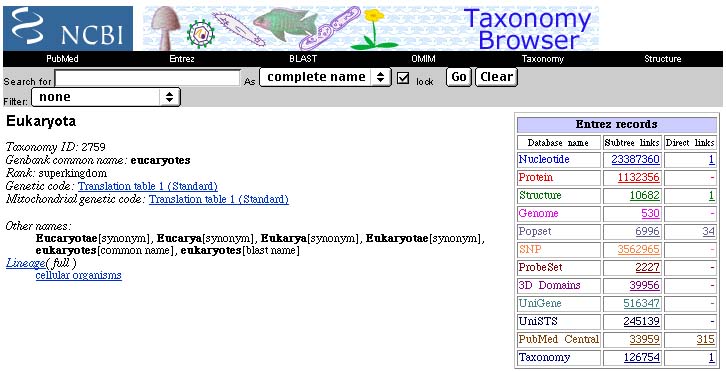
Next (back to July, 2002), I specifically searched for cytochrome c sequences, and came up with about 500 in the protein database. This is not very many at all compared with the thousands of different sequences in the database for some other, larger, protein genes. Even though this includes records for some non-eukaryotes, there are still far more than the 40 eukaryote cytochrome c records claimed in the text. If you would like to look at some 3-D computer generated predicted structures of cytochrome c, click here. To open up a new browser window with an amino acid alignment (from the ProtDom website) of most of the available complete cytochrome c sequences for eukaryotes click here. (The first two sequences are arbitrarily selected bacterial cytochrome c sequences, with names in blue, not yellow.)
RQUE15.2: Click on the last link to view the cytochrome c alignment of amino acids. Scroll down to near the bottom. Can you find the human cytochrome c sequence ("CYC_HUMAN")? Can you find any "shared derived" similarities between the rat ("CYC2_RAT") and mouse ("CYC2_MOUSE") sequences that unite them as rodents relative to other non-rodents? For example, I found that rat and mouse share a valine ("V") residue at position 58 in the alignment, whereas other animals have an isoleucine ("I") at this position. This could be explained by hypothesizing that at some time before rat and mouse last shared a common ancestor, there was a mutation at this position that became fixed in the population, and this is why their common ancestor had a valine, and rat and mouse also have a valine. In other words, isoleucing mutated to valine a single time. Or did it? Scroll up to see if any other organisms have a valine in this position. For example, I found that yeast (a kind of fungus) has a valine in this position. Now we have a problem. A human and even a fruit fly ("CYC_DROME") should be more closely related to a rodent than is a yeast, yet yeast is more similar to the rodents. How is this best explained?
RQUE15.3: This question continues from RQ15.2. Both isoleucine and valine are hydrophobic amino acids, that is, they repel polar molecules such as water, but are attracted to nonpolar molecules such as the lipids in a cell membrane. Can you suggest a plausible hypothesis that might explain why there are two alternative hydrophobic amino acid molecules at this position, instead of a hydrophobic or a hydrophilic amino acid, or do you think it might be a mere coincidence.
RQUE15.4: (Optional) As implied above, the science of reconstructing relationships based on protein sequences has advanced much beyond what is implied in the text. A 1997 article by Sandra Baldauf and Ford Doolittle can be viewed in pdf format here. What sequences is this based on, how many are compared, and about how long is each? What is the take-home point of the paper? Can you find where the human sequence falls in the phylogenetic tree based on these sequences? What is its closest relative in the tree? (Note: Sandra is currently completing much more extensive analyses based on multiple protein-coding genes and more taxa.)
National Academy of Science's Evidence Supporting Biological Evolution
III. Molecular Clock
PBS link: Relationships of Living Species: Calibrating Molecular Clocks (pdf)
PBS link: Molecular Clocks: Proteins That Evolve at Different Rates (pdf)
IV. Hemoglobin
V. Gene Duplication
A gene duplication in New World monkeys
Lecture Notes on Gene Duplications
VI. Evolutionary History of Hemoglobin
American Scientist Article Evoluton of Hemoglobin
Globin Gene Server (see alignment of some mammal hemoglobins here)
Is hemoglobin an exaptation with respect to its adaptive value as an oxygen transport protein?
VII. Pseudogenes
VIII. Homeotic Genes
Please pay special attention to the following PBS link
PBS link: Animal Body Plans and Homeotic Genes
PBS link: Comparative Embryology: The Vertebrate Body (pdf)
IX. Ocular Malformations and Evolution
PBS link: Organs of Extreme Perfection (The Eye)
X. Multilegged Frogs Revisited
What, if anything, is Mitochondrial Eve?
Fossil Hominids: Mitochondrial DNA
Tracking Down Mitochondrial "Eve" and Y Chromosomal "Adam"
Selected Available Articles on Human Evolution
Click link to return to Biology
409 Schedule
or back to Chapter 14
or ahead to Chapter 16
This page created 6/8/02 © D.J. Eernisse, Last Modified 3/27/03, Links Last Completely Checked 3/27/03